Research Article
Romial Joel Ngouénam*
Romial Joel Ngouénam*
Corresponding author
Laboratory of
Microbiology, Department of Microbiology, Faculty of Science, University of
Yaoundé I P.O Box 812, Yaoundé, Cameroon.
E-mail : ndeffothomas@yahoo.fr, Tel: +237 699265636/ 620190046
Bilkissou Njapndounke
Bilkissou Njapndounke
Laboratory of Microbiology, Department of Microbiology, Faculty of Science, University of Yaoundé I P.O Box 812, Yaoundé, Cameroon.
E-mail : njapndounkebilkissou123@yahoo.com
Chancel Hector Momo Kenfack
Chancel Hector Momo Kenfack
Department of nutrition, Food and Bioresource Technology. College of Tecnology, University of Bamenda, PO Box 39 Bambili, Bamenda, Cameroon.
E-mail : momohector@yahoo.fr
Edith Marius Foko Kouam
Edith Marius Foko Kouam
Department of Physiological Sciences and Biochemistry, Faculty of Medicine and Pharmaceutical Sciences, University of Dschang, Cameroon.
E-mail : fokokouam@yahoo.fr
Laverdure Tchamani Piame
Laverdure Tchamani Piame
Research Unit of Biochemistry, Medicinal Plants, Food Science and Nutrition (URBPMAN) Department of Biochemistry, Faculty of Science, University of Dschang, P.O Box 67, Dschang, Cameroon.
E-mail : laverduretchamani@yahoo.com
Ulrich Daquain Fotso Techeu
Ulrich Daquain Fotso Techeu
Research Unit of Biochemistry, Medicinal Plants, Food Science and Nutrition (URBPMAN) Department of Biochemistry, Faculty of Science, University of Dschang, P.O Box 67, Dschang, Cameroon.
E-mail : ulrichedaquain@yahoo.fr
Pierre Marie Kaktcham
Pierre Marie Kaktcham
Research Unit of Biochemistry, Medicinal Plants, Food Science and Nutrition (URBPMAN) Department of Biochemistry, Faculty of Science, University of Dschang, P.O Box 67, Dschang, Cameroon.
E-mail : kapima79@yahoo.fr
François Zambou Ngoufack
François Zambou Ngoufack
Research Unit of Biochemistry, Medicinal Plants, Food Science and Nutrition (URBPMAN) Department of Biochemistry, Faculty of Science, University of Dschang, P.O Box 67, Dschang, Cameroon.
E-mail : fzambou@yahoo.fr.
Abstract
This investigation aimed to evaluate the acidifying, proteolytic, lipolytic and texturizing activities of lactic acid bacteria (LAB) isolated from tropical fruits (pineapple, papaya, orange and banana) collected at Foreke (City of Dschang, Menoua Division, West Cameroon region). Firstly, LAB was isolated and characterized at the generic and technological levels. Secondly, the genetic diversity of LAB isolates was highlighted using Random Amplified Polymorphic DNA (RAPD). As a result, twenty six LAB isolates were facultative heterofermentative and presumptly belonging to the genus Lactobacillus. Only 4 of them (N1B6, N2P4, N3A3, N1O5) showed an acidifying activity higher than 35°D after 6 h of incubation and were considered as isolates of interest. In addition, the highest acidifying activities (47.540 ± 0.505°D, 88.857 ± 0.248 °D, 93.433 ± 0.513 °D) after different incubation times (6, 12 and 24 h) were recorded by N1O5 isolate. All the selected LABs isolates showed growth with proteolysis and the diameter of lysis zones varied from 4.25 ± 0.35 mm (N1B6) to 7.80 ± 0.28 mm (N3A3). Also, lipolytic activity was detected in all LAB with the highest activity (10.05±0.07) highlighted by isolate N1B6. Except for isolate N1O5, all the other isolates produced exopolysaccharides. RAPD showed that isolates N3A3, N1B6 and N2P4 are genetically close and different from isolate N1O5. In addition, all these LAB isolates were different from certain reference LAB strains (Lactiplantibacillus plantarum H21, Lacticaseibacillus paracasei 62L, Lactiplantibacillus paraplantarum 78L, Levilactobacillus brevis 77L) based on RAPD. Thus, given the properties of commercial interest, these isolates can be used to ferment dairy products.
Abstract Keywords
Lactic
acid bacteria, genetic
diversity, commercial
interest, random amplified polymorphic DNA, tropical fruits.
1. Introduction
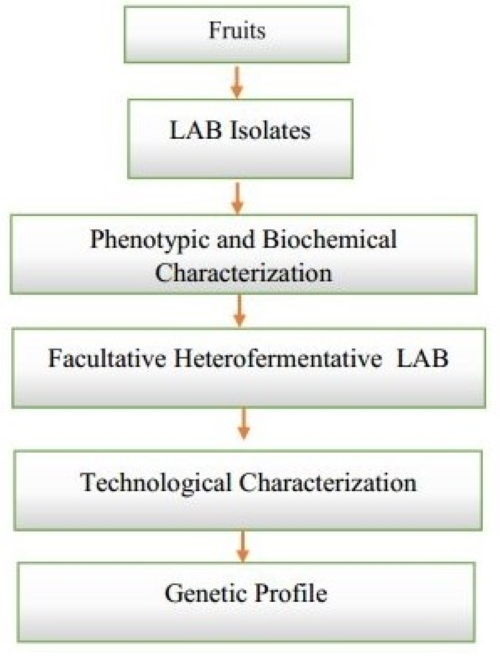
LAB are heterogeneous group of microorganisms that produce lactic acid as the main product of carbohydrate metabolism [1]. Generally recognised as safe, they are increasingly used in the agri-food industries in view of their various technological properties [2]. Indeed, due to their protein and lipid metabolism, they contribute to the development of the organoleptic properties of fermented products through the production of aromatic compounds (α-keto acids, hydroxy acids, aldehydes, ketones, alcohols, carboxylicacids, and esters) [3,4]. Exopolysaccharides (EPS) produced by LAB are responsible for reducing syneresis, increasing viscosity and improving the texture of fermented dairy products. These EPS have also been shown to have beneficial effects on human health (glycemic control, calcium and magnesium absorption, prebiotic, antitumour, antioxidant and antidiabetic, cholesterol reduction, immunomodulatory activities) [5, 6]. One of the most sought-after technological properties in the agri-food industry is antimicrobial activity, which is essential for the safety and preservation of fermented foods. Lactic acid bacteria are very common in nature and occupy a variety of ecological niches. Milk, fermented milk products, meat, digestive tract of animals and traditional fermented drinks have been the subject of isolation of LAB with exploitable technological properties and others used as probiotics [4, 7-11]. Just like the aforementioned niches, tropical fruits (pineapple, papaya, orange and banana) provide a favourable environment for the development of microorganisms [1]. These microorganisms are sometimes responsible for the rapid deterioration of these fruits, and are released into the environment, they contribute to environmental pollution [12]. However, these rotten fruits could be used to isolate LAB with technological properties that can be exploited industrially. Several authors were interested in tropical fruits as an isolation biotope of LAB [1,12-14]. However, none of them have focused on LABs, which have at the same time, acidifying, proteolytic, lipolytic and texturizing activities that could be an advantage in the production of fermented dairy products. In developing countries such as Cameroon, small businesses manufacturing fermented dairy products are expanding rapidly. These entrepreneurs are showing a need for inexpensive local fermenting agents combining the four technological properties aforementioned. These lactic strains would enable them to facilitate fermentation conditions, improve the quality of their products and do so without ordinary lactic ferments, which are expensive and difficult to use.
The present research aimed to evaluate the acidifying,
proteolytic, lipolytic and texturizing activities of LABs isolated from Cameroonian tropical fruits (pineapple, papaya, orange and banana). In
addition, the genetic profile of LAB isolates having all the latter properties was
highlighted.
These LAB isolates would be used as fermentation agents for dairy products.
2.
Materials and methods
2.1. LAB strains and growth conditions
The LAB strains (Table 1) preserved in the reconstituted skim milk (12.5%, (w/v) supplemented with glycerol (25%, v/v) and used in the present work as positive controls for the RAPD were obtained from our microbial bank collection. For each strain, the cultures were obtained after 2 consecutive reactivations (1%, v/v) in the sterilized MRS broth (TM MEDIA, Titan Biotech Ltd, India) at 30 ºC for 18 h.
Table 1. Strains used as positive control for the RAPD
|
LAB strains |
Accession number (NCBI GenBank) |
|
Lactiplantibacillus plantarum H21 |
KU886168 |
|
Lacticaseibacillus paracasei
62L |
KU886178 |
|
Lactiplantibacillus paraplantarum 78L |
KU886183 |
|
Levilactobacillus brevis 77L |
KU886182 |
2.2. LABS isolation and presumptive identification
LABs were isolated by serial dilution plating on MRS medium (TM MEDIA, Titan Biotech Ltd, India) from rotten tropical fruits (orange, banana, papaya and pineapple) collected at Foreke (City of Dschang, Menoua Division, West Cameroon region). All cultures were grown at 30 °C under anaerobic conditions for 48 h. The phenotypic, morphological and biochemical characterization of pure LAB isolates were done as recommended by [15].
2.3. Assessment of the technological
properties of LAB
2.3.1 Acidifying activity
The
method described by Alharbi et al., [2] with
minor modifications was used to carry out the acidifying activity of LAB
isolates. The latter was measured by monitoring the pH of the different
cultures in skimmed milk (10%) after 6, 12 and 24 h of incubation and by measuring
the titratable acidity expressed in Dornic degree (°D).
Only the LAB isolates with the best
acidifying activity after 6 h of incubation were selected as isolates of
interest for further work. After preparing and sterilising the skimmed milk in
100 ml Erlenmeyer flasks, the flasks were cooled and inoculated (1%) with 12
h-old LAB cultures, then incubated at 30 °C for 24 h. Furthermore, 10 ml of milk were taken from the
cultures after different incubation times and the pH was measured directly
using a pH meter (pH meter PT-10, Sartorius AG, Gottingen, Germany). The latter
volume of milk was titrated with sodium hydroxide (0.1 N) in the presence of 2 drops of
phenolphthalein, until the colour changed to pale pink for at least 10 seconds.
The quantitative data obtained were the average of three replications. The titrable
acidity was determined by the following formula :
Where
D: mass in decigram of lactic acid per
litre of milk;
C1: Concentration of the NaOH solution;
Veq: Volume of NaOH used to titrate the
lactic acid contained in the 10 ml of milk;
Vo: Volume of milk used;
M: Molar mass of lactic acid
2.3.2. Proteolytic activity
The proteolytic activity of LAB was evaluated on Plat Count Agar (TM MEDIA, Titan Biotech Ltd, India) medium supplemented with 2% skimmed milk as described by Mechai et al., [16]. After sterilisation, the medium supplemented with skimmed milk was poured and left to solidify. The 12 h-old LAB cultures were deposited as spots on the media and the whole was incubated at the temperatures used to isolate the microorganisms for 48 h. Proteolysis was detected by assessing a clear zone around the colonies. Proteolytic activity was carried out in triplicate.
2.3.3. Lipolytic activity
The modified method of Tanasupawat et al., [17] was used to highlight lipolysis on agar using Tween-20 as the lipid source. The composition of the medium was as follows (per 1000 ml): peptone 10.0g, NaCl 5.0g, CaCl2 0.1 g and agar 15.0 g. Tween-20 (lauric acid ester) was autoclaved separately and added at 1% to the sterile medium previously formulated. The mixture was distributed to the Petri dishes and allowed to solidify. Young cultures (12 h-old cultures) were deposited as spots on the poured media and the whole set was incubated at 30 ºC for 48 h. Lipolysis was detected by the appearance of a clear zone around the spots and a visible precipitate, due to the precipitation of calcium salt crystals containing free fatty acids released by the enzymes produced by the LAB isolates. Lipolytic activity was performed in triplicate.
2.3.4. Investigation of exopolysaccharide production by LAB isolates
The production of exopolysaccharides by LAB isolates was performed using the protocol of Grosu-Tudor, & Zamfir [18]. Young LAB (12 h-old cultures) were streaked on MRS agar supplemented with glucose (20 g/l), lactose (20 g/l) fructose (20 g/l) and sucrose (50 g/l). After 48 h of incubation at 30 ºC, the plates were checked for the presence of large, sticky or mucoid colonies as well as those forming long visible filament as soon as they were touched and lifted by a sterile loop.
2.4. Genetic profiling of isolates of
interest by RAPD
2.4.1 DNA amplification
Because of its reproductivity [19, 20], the minisatellite primer M13 (5'-GAGGGTGGCGGTTCT-3'), was used for the PCR reactions as described by Schillinger et al., [21]. The PCR reaction was carried out in a reaction mixture with a total volume of 25 μL. This reaction mixture consisted of 12.5µl of 2x Master Mix, 0.5 μL of primer, fresh colonies of LAB isolates as well as reference strains (Lactiplantibacillus plantarum H21; Lacticaseibacillus paracasei 62L; Lactiplantibacillus paraplantarum 78L; Levilactobacillus brevis 77L) and 12 μL of sterile ultrapure water. The thermal cycler (Techne 5PRIMEG/02, United Kingdom) was used for this purpose. The reaction mixtures containing the primer M13 were heated to 94 °C for 2 min and then submitted to 30 cycles at 94 °C for 1 min, 42 °C for 20 s, 72 °C for 2 min. At the end of these 30 PCR cycles, the mixtures were left to a final extension period of 10 min at 72 °C to complete any amplifications. A DNA-free amplification (negative control) was carried out in each series to check for any contamination by DNA foreign to the template. The PCR products were then separated by agarose gel electrophoresis.
2.4.2.
Separation of PCR products by agarose gel electrophoresis.
One gram (1 g) of agarose (Molecular Biology Certified) was
dissolved in 100 ml of 1x TAE buffer, boiled and then cooled to 50 °C. Five µl
of ethidium bromide (10 µg/ml) were added. This mixture was then transferred to
the agarose gel pattern (fitted with the well forming comb) and covered for 30 min
to allow gel formation, after which the buffer was introduced onto the gel
surface. For each sample, a mixture of 8 µl of the product and 2 µl of ethidium
bromide was introduced into a well in the gel and the electrophoreograph was
subjected to a voltage of 100 V for 30 min. The DNA bands were visualised and recorded under UV light using the GelDoc2000
Transilluminator (UVITEC, Biorad, Hercules, Canada). The PCR products of the
aforementioned reference strains were used as positifive control. Molecular
weight marker and DNA III Ladder Mix (Tiagen, China) were used to estimate
the different DNA
fragment lengths. Polymorphism was analysed by comparing DNA
bands on the electrophoretic profile.
2.5.
Preservation of LAB
LAB isolates which have at the same time, acidifying, proteolytic, lipolytic and texturizing activities (commercial isolates) were preserved at -20 °C in the reconstituted skim milk (12.5%, (w/v) supplemented with glycerol (25%, v/v) for further needs.
2.6. Statistical analysis
Results were expressed as mean ± standard deviation using Microsoft
Excel version 13 software. Also, the quantitative data obtained in this work
were analysed by the Analysis of Variance (ANOVA) test using Minitab 19
software, followed by comparisons of the means by the Fisher test at 0.05
probability threshold.
3. Results
3.1. Isolation and generic identification
of lactic acid bacteria
The number of LAB isolates obtained from fruit samples (orange, papaya, banana and pineapple) as well as some of their phenotypic and biochemical characteristics are shown in (Table 2).
Table 2. Number and grouping of LAB isolates based on phenotypic and biochemical criteria
|
Biotope Characteristics |
Papaya |
Banana |
Orange |
Pineapple |
||||
|
Number of isolates |
n=12 |
n=14 |
n=26 |
n=9 |
||||
|
Culture medium |
MRS |
MRS |
MRS |
MRS |
||||
|
Morphology |
Rods |
Rods |
Rods |
Rods |
||||
|
Catalase test |
- |
- |
- |
- |
||||
|
Gram staining |
+ |
+ |
+ |
+ |
||||
|
CO2 production |
7-/5+ |
11-/3+ |
4-/22+ |
4-/5+ |
||||
|
Growth at 45°C |
+ |
+ |
+ |
+ |
||||
|
Growth at 10°C |
+ |
+ |
+ |
+ |
||||
|
Genus |
L.g.2 |
L.g.3 |
L.g.2 |
L.g.3 |
L.g.2 |
L.g.3 |
L.g.2 |
L.g.3 |
|
Number |
7 |
5 |
11 |
3 |
4 |
22 |
4 |
5 |
|
Frequency (%) |
58.33 |
41.67 |
78.57 |
21.43 |
15.38 |
84.62 |
44.44 |
55.56 |
|
+ : positif ; - : négatif ; n=number ; L.g.= Lactobacillus group |
||||||||
A total of 61 Gram-positive and catalase-negative LAB isolates were isolated from these samples. Microscopic observation of these isolates revealed that they exhibited rod-like cell shapes with modes of association and sizes varying from one isolate to another. This table also shows that 35 of these isolates produce CO2 from glucose and were therefore classified in the Lactobacillus Group III based on data in Bergey's Manual of Systematic Bacteriology. The remaining 26 LAB isolates were classified in the genus Lactobacillus Group II (facultative heterofermentative lactic acid bacteria) since they grew simultaneously at 45 °C and 10 °C and did not produce CO2 from glucose ; the latter were used for further work.
3.2. Evaluation of the technological properties of homofermentative LAB isolates
3.2.1 Acidifying activity
The acidifying capacity of the isolates, shown by the lowering of the pH and Dornic acidity are shown in Table 3. Of the 26 facultative heterofermentative LAB isolates previously obtained, it emerged that the lactic acid concentration here expressed in degree Dornic varied from one LAB isolate to another. However, only 4 of these isolates stood out from the others after 6 h of incubation with activities greater than 35 °D and were therefore used for the rest of the work. In addition, the acidifying activity of these isolates of interest was also assessed after 8 h and 24 h incubation. The results presented in this table reveal that the highest acidifying activity (47.540 ± 0.505 °D, 88.857 ± 0.248 °D, 93.433 ± 0.513 °D) after different incubation times were recorded by isolate N1O5. Moreover, the aforementioned quantities of lactic acid were significantly different (p < 0.05) from those produced by the other LAB isolates. Our results also showed a low production of lactic acid by LAB isolates between 12 h and 24 h incubation.
Table 3. Time course of acidifying activity of LAB isolates
LAB isolate | Time incubation | |||||
6 h | 12 h | 24 h | ||||
ΔpH (pHt0h–pHt6h) | Acidity (◦D) | ΔpH (pHt0h–pHt12h) | Acidity (◦D) | ΔpH (pHt0h–pHt24h) | Acidity (◦D) | |
N1B6 | 0.820 ± 0.02c | 37.470 ± 0.503c | 1.710±0.010d | 74.433±0.551d | 1.873±0.011d | 80.533±0.503d |
N2P4 | 0.977± 0.030b | 43.610 ±0.535b | 1.890±0.000c | 80.780±0.220c | 1.93±0.010c | 86.27±0.642c |
N3A3 | 0.987± 0.023b | 44.540 ± 0.505b | 1.930±0.010b | 82.97±0.251b | 1.970±0.010b | 88.533±0.472b |
N1O5 | 1.100 ± 0.040a | 47.540 ± 0.505a | 1.960±0.010a | 88.857±0.248a | 2.14±0.050a | 93.433±0.513a |
a,b,c,The values with different letters on the same column differ significantly each other (p < 0.05). N1O5: Lactobacillus isolated from orange; N1B6: Lactobacillus isolated from banana; N2P4: Lactobacillus isolated from papaya; N3A3: Lactobacillus isolated from pineapple; N3A7: Lactobacillus isolated from pineapple. | ||||||
3.2.2 Proteolytic activity
The results of the proteolytic activity of LAB isolates are presented in Table 4. Our results showed that all the isolates had proteolytic activity, highlighted by the appearance of a clear zone around the spots, as shown in Fig. 1. Moreover, the diameters of the lysis zones varied from one isolate to another. N3A3 isolate showed a significantly (p < 0.05) highest inhibition diameter (7.8 mm). However, the smallest proteolytic activity (4.10 mm) was recorded by isolate N1O5, although it was not significantly different from that of isolate N1B6 (p > 0.05).
Table 4. Proteolytic activity of LAB isolates
Isolates | Observation | Diameter of lysis zone (mm) |
N1B6 | Growth with proteolysis | 4.25 ± 0.35a |
N2P4 | Growth with proteolysis | 6.00 ± 0b |
N3A3 | Growth with proteolysis | 7.80 ± 0.28c |
N1O5 | Growth with proteolysis | 4.10 ± 0.14a |
a,b,c, The values with different letters on the same column differ significantly each other (p < 0.05). | ||
3.2.3 Lipolytic activity
Table 5 contains data obtained from the assessment of LAB’s lipolytic activity. The results presented in this table reveal that all the isolates studied showed growth with lipolytic activity, as evidenced by the appearance of a clear zone around the spots and a visible precipitate, formed by the fatty acids released by the enzymes produced by the isolates (Fig. 2). The diameters of the lysis zones vary between 4 and 10.05 mm. Isolate N1B6 recorded the highest activity with a lysis zone diameter of 10.05 mm : a value significantly (p<0.05) different from those of the other isolates.
Figure 2. Clear area around spots marking lipolytic activity of LAB isolates
Table 5. Lipolytic activity of LAB isolates
Isolates | Observation | Diameter of clear zone (mm) |
N1B6 | Growth with lipolysis | 10.05±0.07d |
N2P4 | Growth with lipolysis | 7.90 ± 0.14c |
N3A3 | Growth with lipolysis | 5.90 ± 0.14b |
N1O5 | Growth with lipolysis | 4.00 ±00a |
a,b,c,d The values with different letters on the same column differ significantly each other (p < 0.05). | ||
3.2.4 Texturising activity
The results of the exopolysaccharides production test are shown in Table 6. All LAB isolates, except N1O5, were able to grow on a hypersaccharose medium, forming colonies with a slimy appearance, reflecting the production of a thickening agent, exopolysaccharides. This sliminess is revealed by the formation of a long visible filament when the viscous colonies are touched and lifted with a sterile loop.
Table 6. EPS production by LAB
Isolats | Observation | Test evaluation |
N1B6 | Large, slimy colonies | + |
N3A3 | Large slimy colonies | + |
N2P4 | Large slimy colonies | + |
N1O5 | Normal colonies | - |
3.3 Genetic diversity of LAB isolates by RAPD
Fig. 3 shows the electrophoretic profile of RAPD products using the primer M13 for the best LAB isolates (N1B6, N2P4, N3A3, N1O5) and reference strains (Lactiplantibacillus plantarum H21; Lacticaseibacillus paracasei 62L; Lactiplantibacillus paraplantarum 78L; Levilactobacillus brevis 77L) from the laboratory.
Figure 3. Agarose gel showing electrophoretic profiles of RAPD products ;
M: molecular weight marker; Positive controls: H21, 62L, 78L 77L; CN:
Negative control (ultra pure distilled water).
All the LAB isolates showed electrophoretic profiles that differed from those of the reference strains. This electrophoregram shows that the DNA size of LAB isolates (2000 bp) is smaller than that of reference strains. No DNA fragment was observed in the electrophoretic line of isolate N1O5. However, isolates N1B6, N3A3 and N2P4 have identical electrophoretic profiles because the bands that appeared were at the same migration distance. In addition, the N1B6 band electrophoretic profile was very thick compared to those of the other isolates.
4. Discussion
Bacteria can only develop when they find in their environment the nutrients they need to synthesise their cellular constituents. The basic requirements common to all bacteria are water, a source of energy, nitrogen and vitamins. According to the literature the nutrients mentioned above are found in tropical fruits (banana, pineapple, orange and papaya), so it is not surprising that these are favourable matrices for the growth of microorganisms, particularly for LAB [1, 12, 22]. In the present work, facultative heterofermentative LAB were isolated from tropical fruits ; similar data were reported by several authors who in their investigations on the lactic flora of papaya, guava, pineapple and orange were able to isolate LAB of the genus Lactiplantibacillus plantarum, Lactiplantibacillus pentosus and Levilactobacillus brevis [12,14, 23-25]. LAB can be endowed with technological properties of commercial interest, in particular their acidifying activity.
The ability of LAB to produce lactic acid is of vital importance in the manufacture of fermented dairy products ; as well as having antimicrobial activity, this organic molecule also helps to improve the taste qualities of these dairy products [4, 26]. A fermentation agent with rapid acidifying activity is of great interest, as it reduces product manufacturing time and consequently increases the company's production yield. For these reasons, the selection criterion for the isolates of interest in this work was acidifying activity after 6 h of incubation. In the context of this study, the evolution of acidity and pH variations during the growth of isolates tested in skimmed milk showed a difference between isolates from the four matrices. Boulares et al., [27] reported that this difference could exist between genera, species and even between strains of the same species. The increasing acidity in the sterile milk over time is thought to result from the breakdown of lactose by LAB to produce the energy required for the synthesis of their constituents and their growth, while producing lactic acid, hence the increase in acidity in the environment. This acidification would consequently lead to a lowering pH of the medium; these observations are in line with data in the literature [2, 28]. In the present work, the slight acidification observed after 12 h of incubation can be explained by the significant reduction in the quantity of carbohydrate substrate in the medium, but also by retro-inhibition due to the accumulation of lactic acid [5]. The quantities of lactic acid produced (37.470 - 47.540 °D) by the LAB isolates of interest (N1B6, N2P4, N3A3, N1O5) after 6 h of incubation are higher than those obtained by Alharbi et al., [2]. In fact, the acidity values obtained by these authors whose work focused on the characterisation of LAB isolated from raw milk in Saudi Arabia were between 20.5 and 27.3 °D after 6 h of incubation. Furthermore, the highest concentration of LA (9.343 g/l) obtained in this study after 24 h of incubation is lower than the highest (10.43 g/l) concentration recorded by Alharbi et al., [2] in their work at the same incubation time. This difference could be explained by the fact that the acidifying activity in the milk depends not only on the lactic ferment, but also on its isolation biotope, sampling area and fermentation conditions (initial carbohydrate concentration) [28-30]. The growth of LAB isolates is closely linked to their ability to degrade protein (casein) present in the milk as nitrogen source.
The proteolytic activity of LAB is essential for their growth in protein-rich substrates and for the development of the organoleptic properties of fermented products through the production of aromatic compounds resulting from the catabolism of amino acids (α-keto acids, hydroxy acids, aldehydes, ketones, alcohols, carboxylicacids, and esters) [4]. Bettache et al. [30] showed that lactobacilli isolated from Dhan, an Algerian fermented milk product, exhibited proteolytic activity on PCA medium with 2% skimmed milk. In their investigations, Abubakr et al., [13] showed that LAB from banana, date, apple and pineapple in India were proteolytic and that the lysis zones observed varied between isolates from different fruits and even between those from the same biotope. Similar data have been highlighted within the framework of the present investigation. Just like proteins, lipid degradation by LAB is of vital importance in the production of fermented dairy products.
Béal et al., [3], in their studies on the production and conservation of lactic ferments and probiotics, showed that lactobacilli have lipolytic activity and are therefore of interest for certain cheese applications. In the context of the present investigation, the lipolytic activity tested on tween 20 agar medium was pronounced in all four (4) LAB isolates of interest, with significant zones of lysis varying from one isolate to another. These results are in line with those of Bettache et al., [31] who showed that LAB of the genus Lactobacillus (Lactobacillus delbrueckii subsp. delbrueckii, Lactobacillus delbrueckii subsp. Bulgaricus) from an Algerian dairy product named Dhan, showed lipolytic activity on agar medium containing tween 20 (lauric acid ester) and that the lysis zones varied from one strain to another. The same applies to the work of Alharbi et al., [2] who highlighted the expression of extracellular lipolytic activity in LAB isolated from raw milk in Saudi Arabia.
This research also aimed to evaluate the ability of LAB to produce EPS. The production of EPS by lactic bacteria is a phenomenon that is favourable to a number of industrial food processes (reducing syneresis, increasing product viscosity and improving texture), particularly in the manufacture of fermented dairy products [12, 32]. EPS have also been shown to have health benefits in humans (glycemic control, calcium and magnesium absorption, prebiotic, antitumour, antioxidant and antidiabetic, cholesterol reduction and immunomodulatory activities) [5,6, 33]. The texturising activity was found in 3 isolates (N1B6, N2P4, N3A3), which showed an ability to form large, sticky colonies indicating EPS production. Similar observations have been reported by Georgieva et al., [5] who showed in their studies on the isolation and selection of Sauerkraut LAB producing exopolysaccharides that out of a collection of 20 LAB isolates, only 6 were capable of producing EPS. This observation could be explained by the non expression of the genes involved in the synthesis of the enzymes responsible for the production of EPS in LAB as reported by Meulen et al., [34]. The molecular identification of LAB with exploitable technological properties is an essential requirement in microbiology.
The RAPD technique was first described in 1990 [35], and has been used to study the dynamics of microorganisms responsible for food fermentation and to differentiate Lactobacillus strains [20]. In this study, the use of the RAPD technique was applied using the M13 primer to detect genotypic differences between the isolates. The absence of an amplification band as shown on the electrophoretic profile for isolate N1O5 suggests that the M13 primer used did not find a targeted DNA sequence in its genome. Another explanation would be the non-release of bacterial genetic material during PCR due to the absence of heat-induced rupture of the bacterial wall. Isolates N1B6, N3A3 and N2P4 showed identical electrophoretic profiles (same length and same migration fronts), which would probably mean that they are genetically close, and genetically different from the isolate N1O5. Similar data were reported by Schillinger et al., [21] whose work focused on the use of group-specific and RAPD analyses for rapid differentiation of Lactobacillus strains from probiotic yogurts. The thickness of the band that appears on the electrophoretic profile of isolate N1B6 would be due to the DNA concentration which was very high at this level. PCR products of the four (4) aforementioned LAB isolates were different in size from those (electrophoretic profiles marked by the absence or presence of bands with different migration distances) of the reference species. This observation can be explained by the fact that these isolates were genetically different from the reference species. These results are in agreement with those of Zapparoli et al., [19] who demonstrated in the course of their work on LAB differentiation by randomly amplified polymorphic DNA and pulsed-field gel electrophoresis that the Lactobacillus sanfranciscensis strains with the same electrophoretic profile were genetically close. According to the previous explanations, the N1O5, N1B6, N3A3 and N2P4 LAB isolates obtained during this work would not be genetically close to the species Lactiplantibacillus plantarum H21, Lacticaseibacillus paracasei 62L, Lactiplantibacillus paraplantarum 78L, Levilactobacillus brevis 77L which were used as reference strains during RAPD. However, to remove this doubt, molecular identification of the LAB using 16S rRNA gene sequencing will be required.
5. Conclusions
Based on the present research, three, facultative heterofermentative LAB (N1B6, N2P4 and N3A3) isolates having at the same time acidifying, proteolytic, lipolytic and texturising properties were obtained from tropical fruits (pineapple, papaya and banana). However, these properties were of varried degrees. Among these isolates, N3A3, N1B6 and N2P4 were genetically close and both were different from isolate N1O5. Concerning the molecular identification of LAB isolates, the RAPD used for this purpose did not make it effective. Indeed, all the LAB isolates were different from the reference strains (Lactiplantibacillus plantarum H2; Lacticaseibacillus paracasei 62L; Lactiplantibacillus paraplantarum 78L; Levilactobacillus brevis 77L). Hence, given the commercial interest properties of these LAB, they could have applications in the agri-food industry. To this end, further work would be aimed at evaluating the safety properties and performing the molecular identification of the LAB isolate using 16S rRNA gene sequencing.
Authors’ contributions
Designed, contributed and carried out the study, data analysis, interpreted the results and drafted the article, R.J.N.; Analyzed and interpreted data, B.N., C.H.M.K., E.M.F.K., L.T.P., U.D.F.T. and P.M.K.; Conceived and designed the experiments, contributed to the arrangement of reagents, materials and data analysis tools. Contributed to drafting of the article, F.Z.N.
Acknowledgements
The authors are grateful to Ulrich Landry Bemmo Kamdem, Jules‑Bocamdé Temgoua, Serge Cyrille Houketchang Ndomou, for their help during the manipulations.
Funding
This research study didn’t receive any specific grant from public, commercial funding agencies or not-for-profit sectors.
Availability of data and materials
All data will be made available on request according to the journal policy
Conflicts of interest
The authors declare no conflict of interest
References
1.
Rodríguez, L.G.; Mohamed, F.;
Bleckwedel, J.; Medina, R.; De Vuyst, L.; Hebert, E.M.; Fernanda M.F. Diversity and
functional properties of lactic acid bacteria isolated
from wild fruits and flowers present in northern Argentina. Front. Microbiol. 2019, 10, Article 1091.
http://dx.doi.org/10.3389/fmicb.2019.01091.
2.
Alharbi, N.k.; Alsaloom N.A.
Characterization of lactic bacteria isolated from raw milk and their
antibacterial activity against bacteria as the cause of clinical bovine
mastitis. J.
Food Qual. 2022, 2022, 1-8.
http://dx.doi.org/10.1155/2022/9854598.
3.
Béal, C.; Marin, M.; Fontaine, E.;
Fonseca, F.; Obert, J. Production
et conservation des ferments lactiques et probiotiques. In Bactéries lactiques, de la génétique aux ferments (p 848). Editions
Lavoisier. Paris, France : Tec
& Doc-Lavoisier, 2008.
4.
Zareie, Z., Moayedi, A.; Garavand,
F.; Tabar-Heydar, K.; Khomeiri, M.; Maghsoudlou, Y. Probiotic properties,
safety assessment, and aroma-generating attributes of some lactic acid bacteria
isolated from Iranian traditional cheese. Fermentation. 2023, 9(4), 338. http://dx.doi.org/10.3390/fermentation9040338.
5.
Georgieva, A.; Petkova, M.; Todorova, E.; Gotcheva, V.;
Angelov, A. Isolation and selection of
sauerkraut lactic acid bacteria producing exopolysaccharides. BIO Web Conf. 2023, 58, 1-6. http://dx.doi.org/10.1051/bioconf/20235802001.
6.
Jurášková, D.; Ribeiro, S.C.; Silva, C.C. Exopolysaccharides produced by
lactic acid bacteria: from biosynthesis to health-promoting properties. Foods.
2022, 11(2), 1-23. http://dx.doi.org/10.3390/foods11020156
7.
Bemmo, K.U.; Kaktcham P.M.; Momo K.C,; Foko, K.E.; Zambou
N.F.; Y, W.R.; Zhu, T.; Li, Y. Bile salt
hydrolase and antimicrobial activities of three bile resistant probiotics Lactobacillus plantarum strains isolated
from Cameroonian artisanal fermented milk. J. Microbiol. Biotechnol. Res. 2017,
7(5), 21-30. http://dx.doi.org/10.24896/jmbr.2017754.
8.
Foko, K.E.; Zambou, N.F.; Kaktcham, P.M.; Wang R.Y.; Zhu T.;
Yin L. Screening and characterization of Lactobacillus
sp. from the water of Cassava’s fermentation for selection as probiotics. Food
Biotechnol. 2018, 32(1), 15-34. http://dx.doi.org/10.1080/08905436.2017.1413984.
9.
Fotso, T.U.; Kaktcham, P.M.;
Kenfack M.H.; Foko K.E.; Tchamani P.L.; Ngouenam, R.J.; Zambou, N.F.
Isolation, characterisation, and effect on biofilm formation of bacteriocin produced by Lactococcus lactis F01 isolated from Cyprinus carpio and application for biopreservation of fish sausage.
Biomed Res. Int. 2022, 2022, 1-15. http://dx.doi.org/10.1155/2022/8437926.
10.
Kaktcham, P.M.; Tchamani, P.L.; Sandjong, S.G.; Foko, K.E.;
Temgoua, J.; Zambou, N.F.; Pérez‑Chabela, M.L. Bacteriocinogenic Lactococcus lactis subsp. lactis 3MT isolated from freshwater Nile
Tilapia: isolation, safety traits, bacteriocin characterisation, and application
for biopreservation in fsh pâté. Arch. Microbiol. 2019, 201(9), 1-10. http://dx.doi.org/10.1007/s00203-019-01690-4.
11.
Momo, C.H.; Zambou, N.F.; Kaktcham, P.M.;Wang R.Y.; Zhu T.; Yin, L. Safety and
antioxidant properties of five probiotic Lactobacillus
plantarum strains isolated from the digestive tract of honey Bees. Am. J. Microbiol.
Res. 2018, 6(1), 1-8.
http://dx.doi.org/10.12691/ajmr-6-1-1.
12.
Ngouénam, R.J.; Momo, K.C.; Foko, K.E.; Kaktcham, P.M.;
Rukesh, M.; Zambou, N.F. Lactic acid production ability of Lactobacillus sp. from four tropical fruits using their by-products
as carbon source. Heliyon. 2021a. 7(5), e07079.
http://dx.doi.org/10.1016/j.heliyon.2021.e07079.
13.
Abubakr, M.A.; Hassan, Z.; Imdakim, M.M.; Sharifah, N.R.
Antioxidant activity of lactic acid bacteria (LAB) fermented skim milk as
determined by 1, 1-diphenyl-2-picrylhydrazy (DDPH) and ferrous chelating
activity (FCA). Afr.
J. Microbiol. Res. 2012, 6(34),
6358-6364. http://dx.doi.org/10.5897/ajmr12.702.
14.
Maryam, A.; Wedad, M. Isolation and identification of lactic acid
bacteria from different fruits with proteolytic activity. Int. J. Microbiol. Biotechnol.
2017, 2(2), 58-64.
15. De
Vos, P.; Garrity, G.M.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.;
Schleifer K.; Withman W.B. Bergey’s Manual of Systemaric Bacteriology (2nd ed). New
york, USA: Springer, 2009.
16.
Mechai, A.; Debabza, M.; Kirane, D. Screening of
technological and probiotic properties of lactic acid bacteria isolated from
Algerian traditional fermented milk products. Int. Food Res. J. 2014, 21(6), 2451-2457.
17.
Tanasupawat, S.; Phoottosavako, M.; & Keeratipibul, S. Characterization and
lipolytic activity of lactic acid bacteria isolated from Thai fermented meat. J. Appl. Pharm.
Sci. 2015, 5(03), 006-012. http://dx.doi.org/10.7324/japs.2015.50302.
18.
Grosu-Tudor, S.; Zamfir, M. Exopolysaccharide
production by selected lactic acid bacteria isolated from fermented vegetables.
Food
Biotechnol. 2014, 18, 107-114.
19.
Zapparoli, G.; Torriani, S.; Dellaglio, F. Differentiation of Lactobacillus
sanfranciscensis strains byrandomly amplified polymorphic DNA and
pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 1998, 166(2), 325-332.
http://dx.doi.org/10.1111/j.1574-6968.1998.tb13908.x.
20.
Sieladie, D.V.; Zambou, N.F.; Kaktcham, P.M.; Cresci,
A.; Fonteh, F.
Probiotic properties of lactobacilli strains isolated from raw cow milk in the
western highlands of Cameroon. Innov. Rom. Food Biotechnol. 2011, 9, 12-28.
21.
Schillinger, U.; Yousif,
N.M.; Sesar, L.; Franz, C.M. Use of groupspecific and RAPD-PCR analyses
for rapid differentiation of Lactobacillus strains from probiotic yogurts. Curr. Microbiol. (2003). 47(6),
453-456. http://dx.doi.org/10.1007/s00284-003-4067-8.
22.
Feumba, D.; Ashwini, R.; Ragu, S. Chemical composition
of some selected fruit peels. Eur. J. Food Sci. Technol. 2016, 4(4), 12-21.
23.
Maria, P. Metabolic engineering of lactic acid bacteria for
the production of industrially important compounds. Comput Struct Biotechnol J.
2012, 3(4), 1-8. http://dx.doi.org/10.5936/csbj.201210003.
24.
Pundir, R.K. Probiotic potential of lactic acid
bacteria isolated from food samples: an in vitro study. J. Appl. Pharm. Sci. 2013. http://dx.doi.org/10.7324/japs.2013.30317.
25.
Todorov, S.; Prévost, M.; Dousset, X.; Le Blanc, J.; Franco, B. Bacteriocinogenic Lactobacillus plantarum ST16Pa isolated
from papaya (Carica papaya)- from
isolation to application: Characterization of a bacteriocin. Int. Food Res. 2011, 44(5),
1351-136. http://dx.doi.org/10.1016/j.foodres.2011.01.027.
26. Ngouénam, R.J.; Kaktcham, P.M.; Momo,
K.C.; Foko, K.E.; Zambou, N.F. Optimization of lactic acid production from pineapple
by-products and an inexpensive nitrogen source using Lactiplantibacillus plantarum strain 4O8. Int. J. Food Sci. 2021b. 2021, 1-11. http://dx.doi.org/10.1155/2021/1742018.
27.
Boulares, M.; Aouadhi, C.; Melika, M.; Moussa, O.B.; Essid,
I.; Hassouna, M. Characterisation, identification and technological properties
of psychotrophic lactic acid bacteria originating from tunisian fresh fish. J.
Food Saf. 2012, 32(3), 333-344. http://dx.doi.org/10.1111/j.1745-4565.2012.00385.x.
28. Abarquero, D.; Renes, E.;
Combarros-Fuertes, P.; Fresno, J.M.; Tornadijo, M.E. Evaluation of
technological properties and selection of wild lactic acid bacteria for starter
culture development. Lwt-food. Sci. Technol. 2022, 171. http://dx.doi.org/10.1016/j.lwt.2022.114121.
29.
Amani, E.; Eskandari1, M.H.;
Shekarforoush, S. The effect of proteolyti activity of starter cultures on
technologically important properties of yogurt. Food Sci Nutr. 2017, 5(3), 525-537. http://dx.doi.org/10.1002/fsn3.427.
30.
Faria, A.S.; Fernandes, N.;
Cadavez, V.; Gonzales-Barron, U.
Screening of lactic acid bacteria isolated from artisanally produced alheira
fermented sausages as potential starter cultures. Foods. 2021, 68, 1-6.
31.
Bettache, G.; Fatma, A.; Miloud,
H.; Mebrouk, K. Isolation
and identification of lactic acid bacteria from dhan, a traditional butter and
their major technological traits. World Appl. Sci. J. 2012, 17(4), 480-488.
32.
Berthold-Pluta, A.B.; Pluta1,
A.S.; Garbowska M.; Stasiak-Różańska, L. Exopolysaccharide-producing lactic
acid bacteria– health-promoting properties and application in the dairy
industry. Adv Microbiol. 2019,
58(2), 191-204. http://dx.doi.org/10.21307/pm-2019.58.2.191.
33.
Ma'unatin, A.; Harijono, H.; Zubaidah, E.; Muhaimin, R.M. The
isolation of exopolysaccharide-producing lactic acid bacteria from lontar (Borassus
flabellifer L.) sap. Iran. J. Microbiol.
2020, 12(5), 437-444. http://dx.doi.org/10.18502/ijm.v12i5.4605.
34.
Meulen, R.V.; Grosu-Tudor, S.; Mozzi, F.; Vaningelgem, F.;
Zamfir, M.; De Valdez, G.F.; De Vuyst, L. Screening of lactic
acid bacteria isolates from dairy and cereal products for exopolysaccharide
production and genes involved. Int. J. Food Microbiol. 2007, 118(3), 250-258.
http://dx.doi.org/10.1016/j.ijfoodmicro.2007.07.014.
35.
Cœuret, V.; Dubernet, S.; Bernadeau M.; Gueguen, M.; Vernoux, J.P. Isolation, characterization and
identification of lactobacilli focusing mainly on cheese and other dairy
products. Lait.
2003, 83(4), 269-306. https://doi.org/10.1051/lait:2003019.
This work is licensed under the
Creative Commons Attribution
4.0
License (CC BY-NC 4.0).
Abstract
This investigation aimed to evaluate the acidifying, proteolytic, lipolytic and texturizing activities of lactic acid bacteria (LAB) isolated from tropical fruits (pineapple, papaya, orange and banana) collected at Foreke (City of Dschang, Menoua Division, West Cameroon region). Firstly, LAB was isolated and characterized at the generic and technological levels. Secondly, the genetic diversity of LAB isolates was highlighted using Random Amplified Polymorphic DNA (RAPD). As a result, twenty six LAB isolates were facultative heterofermentative and presumptly belonging to the genus Lactobacillus. Only 4 of them (N1B6, N2P4, N3A3, N1O5) showed an acidifying activity higher than 35°D after 6 h of incubation and were considered as isolates of interest. In addition, the highest acidifying activities (47.540 ± 0.505°D, 88.857 ± 0.248 °D, 93.433 ± 0.513 °D) after different incubation times (6, 12 and 24 h) were recorded by N1O5 isolate. All the selected LABs isolates showed growth with proteolysis and the diameter of lysis zones varied from 4.25 ± 0.35 mm (N1B6) to 7.80 ± 0.28 mm (N3A3). Also, lipolytic activity was detected in all LAB with the highest activity (10.05±0.07) highlighted by isolate N1B6. Except for isolate N1O5, all the other isolates produced exopolysaccharides. RAPD showed that isolates N3A3, N1B6 and N2P4 are genetically close and different from isolate N1O5. In addition, all these LAB isolates were different from certain reference LAB strains (Lactiplantibacillus plantarum H21, Lacticaseibacillus paracasei 62L, Lactiplantibacillus paraplantarum 78L, Levilactobacillus brevis 77L) based on RAPD. Thus, given the properties of commercial interest, these isolates can be used to ferment dairy products.
Abstract Keywords
Lactic
acid bacteria, genetic
diversity, commercial
interest, random amplified polymorphic DNA, tropical fruits.
This work is licensed under the
Creative Commons Attribution
4.0
License (CC BY-NC 4.0).
This work is licensed under the
Creative Commons Attribution 4.0
License.(CC BY-NC 4.0).



